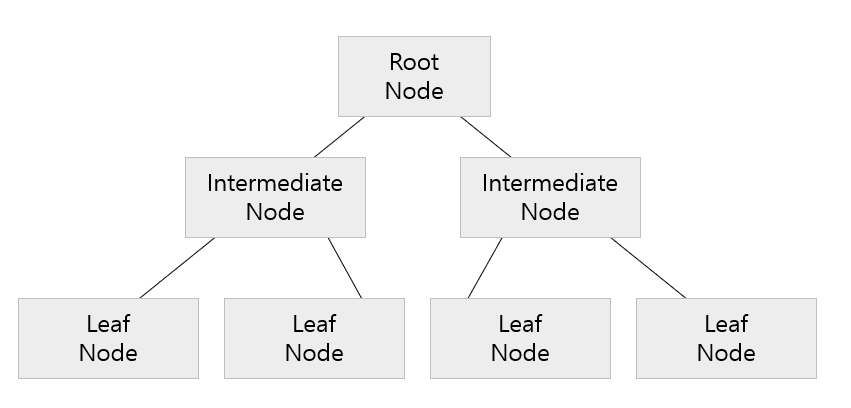
[개념]
- Root Node에서 LeafNode까지 특정한 기준에 맞는지/맞지 않는지를 데이터를 분류한다.
- Node에서 데이터를 분류하는 기준은 2개 이상의 데이터 타입이 서로 얼마나 섞이지 않았고 잘 구분하는지(Impurity :불순도)로 정해진다.
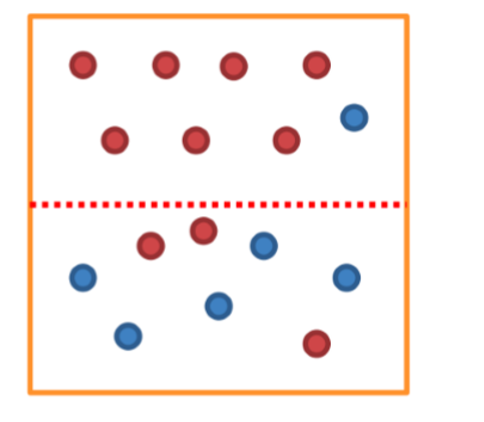
- 위 이미지에서 점선을 기준으로 위쪽으로는 빨간 공이 7/8, 점선 아래로는 파란 공이 4/7로 분류한다.
- 의사결정나무는 추가적으로 점선을 그어 빨간공과 파란공이 섞이는 정도(impurity)를 최소화하는 것을 목표로 한다.
[가지치기]
- 의사결정나무의 깊이가 깊어지면 더 많은 기준을 사용하여 정교하게 데이터를 분류할 수 있으나(impurity↓),
이는 사용 중인 데이터에 대한 의존성이 높아져 Overfitting 문제를 야기할 수 있다.
- 따라서 impurity과 overfitting을 낮추기 위해 의사결정나무의 깊이를 제한해주어야 하는데, 이를 가지치기(pruning)이라고 한다.
★overfitting(과적합)
: 모델이 학습 데이터의 impurity를 높이기 위한 과정에서 학습 데이터에만 지나치게 잘 맞게 학습하여 테스트 데이터에서 오류가 증가하는 문제가 발생
[코드]
1) 데이터 불러오기
2) 데이터 파악
3) Training set(학습 데이터)과 Test set(테스트 데이터)를 분리
4) 모델 생성 및 학습
5) 예측
6) 평가
1) 데이터 불러오기
import numpy as np
import pandas as pd
from pandas import DataFrame
import matplotlib.pyplot as plt
from sklearn import datasets
import seaborn as sns
cancer=load_breast_cancer()
2) 데이터 파악
print("1. cancer.keys : \n{}".format(cancer.keys()))
'''
# 1. cancer.keys : cancer의 함수와 속성 출력(=dir(cancer))
dict_keys(['data', 'target', 'frame', 'target_names', 'DESCR', 'feature_names', 'filename'])
'''
print("2. shape of Cancer : \n{}".format(cancer.data.shape))
'''
# 2. shape of Cancer : cancer 데이터의 행열 개수
(569, 30)
''
print("3. Sample Counts per class : \n{}".format(
{n:v for n,v in zip(cancer.target_names,np.bincount(cancer.target))}
))
'''
# 3. Sample Counts per class : class 별 데이터의 개수
{'malignant': 212, 'benign': 357}
'''
print("4. Feature Names : \n{}".format(cancer.feature_names))
'''
# 4. Feature Names : cancer 데이터의 특징(데이터프레임에서 컬럼명에 해당)
['mean radius' 'mean texture' 'mean perimeter' 'mean area'
'mean smoothness' 'mean compactness' 'mean concavity'
'mean concave points' 'mean symmetry' 'mean fractal dimension'
'radius error' 'texture error' 'perimeter error' 'area error'
'smoothness error' 'compactness error' 'concavity error'
'concave points error' 'symmetry error' 'fractal dimension error'
'worst radius' 'worst texture' 'worst perimeter' 'worst area'
'worst smoothness' 'worst compactness' 'worst concavity'
'worst concave points' 'worst symmetry' 'worst fractal dimension']
'''
dir(cancer)
# dir() : cancer 객체가 가진 속성 또는 함수를 출력
# ['DESCR', 'data', 'feature_names', 'filename', 'frame', 'target', 'target_names']
np.bincount(cancer.target)
# target인 악성/양성에 해당하는 데이터의 개수를 나타냄
# [212 357] : 악성 212, 양성 357개
cancer.DESCR
# 아래와 같이 cancer 데이터셋에 대한 정보를 출력
'''
Breast cancer wisconsin (diagnostic) dataset
--------------------------------------------
**Data Set Characteristics:**
:Number of Instances: 569
:Number of Attributes: 30 numeric, predictive attributes and the class
:Attribute Information:
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three
worst/largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 0 is Mean Radius, field
10 is Radius SE, field 20 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
:Summary Statistics:
===================================== ====== ======
Min Max
===================================== ====== ======
radius (mean): 6.981 28.11
texture (mean): 9.71 39.28
perimeter (mean): 43.79 188.5
area (mean): 143.5 2501.0
smoothness (mean): 0.053 0.163
compactness (mean): 0.019 0.345
concavity (mean): 0.0 0.427
concave points (mean): 0.0 0.201
symmetry (mean): 0.106 0.304
fractal dimension (mean): 0.05 0.097
radius (standard error): 0.112 2.873
texture (standard error): 0.36 4.885
perimeter (standard error): 0.757 21.98
area (standard error): 6.802 542.2
smoothness (standard error): 0.002 0.031
compactness (standard error): 0.002 0.135
concavity (standard error): 0.0 0.396
concave points (standard error): 0.0 0.053
symmetry (standard error): 0.008 0.079
fractal dimension (standard error): 0.001 0.03
radius (worst): 7.93 36.04
texture (worst): 12.02 49.54
perimeter (worst): 50.41 251.2
area (worst): 185.2 4254.0
smoothness (worst): 0.071 0.223
compactness (worst): 0.027 1.058
concavity (worst): 0.0 1.252
concave points (worst): 0.0 0.291
symmetry (worst): 0.156 0.664
fractal dimension (worst): 0.055 0.208
===================================== ====== ======
:Missing Attribute Values: None
:Class Distribution: 212 - Malignant, 357 - Benign
:Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian
:Donor: Nick Street
:Date: November, 1995
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets.
https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needle
aspirate (FNA) of a breast mass. They describe
characteristics of the cell nuclei present in the image.
Separating plane described above was obtained using
Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree
Construction Via Linear Programming." Proceedings of the 4th
Midwest Artificial Intelligence and Cognitive Science Society,
pp. 97-101, 1992], a classification method which uses linear
programming to construct a decision tree. Relevant features
were selected using an exhaustive search in the space of 1-4
features and 1-3 separating planes.
The actual linear program used to obtain the separating plane
in the 3-dimensional space is that described in:
[K. P. Bennett and O. L. Mangasarian: "Robust Linear
Programming Discrimination of Two Linearly Inseparable Sets",
Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu
cd math-prog/cpo-dataset/machine-learn/WDBC/
.. topic:: References
- W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction
for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on
Electronic Imaging: Science and Technology, volume 1905, pages 861-870,
San Jose, CA, 1993.
- O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and
prognosis via linear programming. Operations Research, 43(4), pages 570-577,
July-August 1995.
- W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques
to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994)
163-171.
'''
3) Training set(학습 데이터)과 Test set(테스트 데이터)를 분리
# cancer 데이터의 속성값을 할당
X=cancer.data
# cancer 데이터의 라벨 할당
y=cancer.target
# train과 test set으로 분리
X_train,X_test,y_train,y_test=train_test_split(X,y,test_size=0.2)
4) 모델 생성 및 학습
# DecisionTreeClassifier()모델 적용
dTree=DecisionTreeClassifier()
# fit함수fh DecisionTree모델에 trainset 학습
dTree.fit(X_train,y_train)
5) 예측
# predict 함수에 test set을 넣으면 라벨값을 예측한다.
pred=dTree.predict(X_test)
6) 평가
# accuracy_score import
from sklearn.metrics import accuracy_score
# accuracy_score(테스트 라벨,예측값,옵션)
# normalize=True : 맞춘 확률, False라고 지정하면 맞은 개수
# # dTree.score(X_test,y_test)와 같은 기능
score=accuracy_score(y_test,pred,normalize=True)
score # 0.9298245614035088
- 트리구조 시각화
# graphviz import하기 전에 설치 필요
import graphviz
from sklearn.tree import export_graphviz
export_graphviz(dTree,out_file="dTree.txt", # 사용할 데이터, 데이터를 txt파일로 저장
class_names=cancer.target_names, # class명
feature_names=cancer.feature_names, # feature명
filled=True, # 노드에 색깔주기
leaves_parallel=True, # 리프노드를 기준으로 평평하게
impurity=True) # 불순도(gini) 표시 (True가 기본값)
with open("dTree.txt",encoding="utf-8") as f: # 이미지로 내보내기
dot_graph=f.read();
dot=graphviz.Source(dot_graph)
display(dot)
dot.render(filename="dTree.pdf")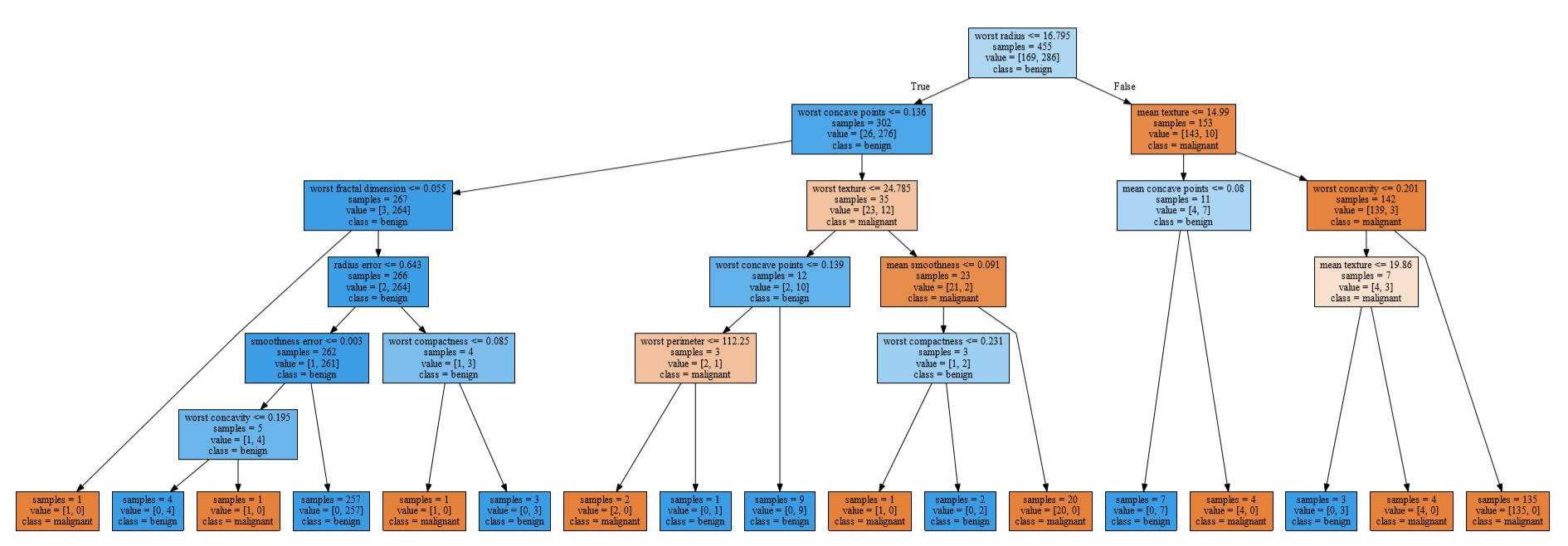
'Data_Analysis > MachineLearning' 카테고리의 다른 글
| [Machine Learning] Kernel SVM(Support Vector Machine) (0) | 2020.12.28 |
|---|---|
| [Machine Learning] SVM(Support Vector Machine) (0) | 2020.12.28 |
| [Machine Learning] NLP - 텍스트처리(Tokenize) (0) | 2020.10.31 |
| [Machine Learning] Ensemble - Random Forest (0) | 2020.07.24 |



댓글